Pedigrees, Family Trees, Autosomal DNA, and Me
by Kyle Davenport, 2012
I grew up with an extended family , and heard talk of relatives who had passed on. My mom had a large typewritten genealogy about her mom's mom's family - the Gage's. It went back to the 17th century and had thousands of descendants of the patriarch William Gage of Freetown , Mass. I thought it was fascinating but couldn't believe anyone had gone to so much work to collect all that information in the 1920's and 1930's. Oh how laborious it must have been to visit all those libraries and court houses across the country, and contact by mail all those descendants!
I thought I would take a stab at my own family tree for a school project when I was 16. Mom's side was easy with the Gage genealogy in hand, and my grandfather told me about his family in southern Indiana. I wrote my dad's mom for any information on that side of the family. She carefully laid out a tree with all the names she knew about – back 3 generations, and told me many interesting stories about them. She didn't know as much about my dad's dad, who had already passed away. “George Davenport was born in Jackson Co, Tenn” was all I had to go on there. (George was my ggf) I even found some information about my ancestors in the library and sent mail to various court houses. For my report I could include lots of old photos and made a chart going back to most of my great-great-grandparents. I talked about life back then and how they migrated with the frontier. They were all humble farmers on the American frontier, and pioneers to unsettled lands.
Well, after the A+ on my school project, the genealogy lay undisturbed for 30 years, until my mom died. As we cleared out her house, I found that genealogy and many more old letters and photos. I decided to piece together all this information into a new genealogy using software and the internet. I missed dear old mom, and doing this genealogy felt like I was reconnecting with her – remembering her and all the others that had gone on before. In the Eighties, a cousin had done a great job collecting information on our shared line, my mom's fathers'. I also inherited all the old photos and letters from my dad's mom. When I put these all together, I had a great tree with dozens of ancestors staring back at me.
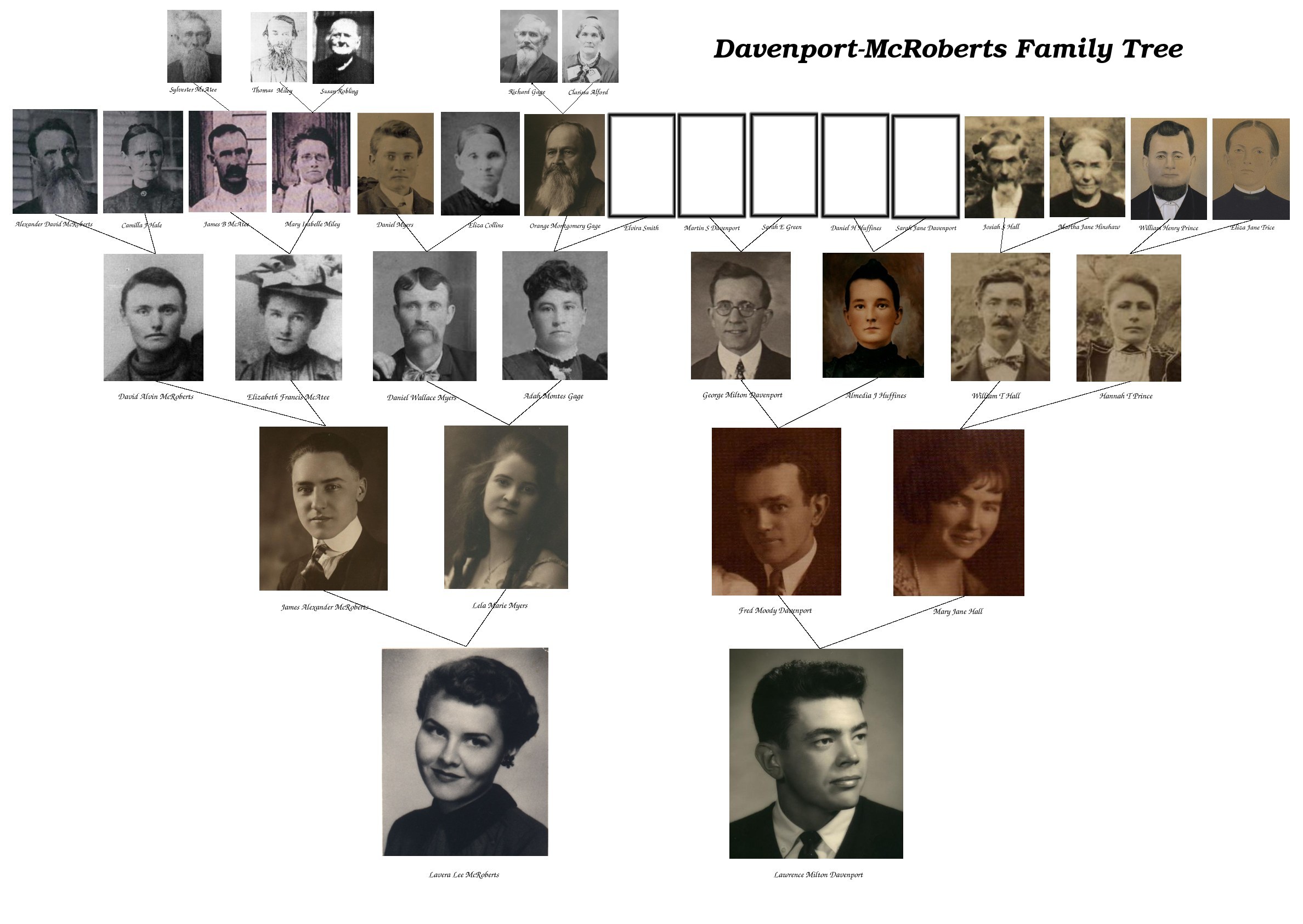
But genealogy on the internet had arrived. I discovered most of my great-great-grandparents in other trees on Rootsweb's WorldConnect ( one good rule of thumb – “If you can push your tree back to great-great-grandparents, it's likely you will find them in someone else's tree”). My elusive Davenports were right there on TN USGenWeb and talked about in GenForum's forums. With ancestry.com, I could find the original references and do my own research too. I started contacting distant cousins I hadn't known before. (Another rule of thumb – “About 1 in 200 of your distant cousins will also be amateur genealogists”) Some of those distant relatives even had photos of my own ancestors which I didn't have, so we were all sharing our information with each other – again, made very easy by the internet.
On WorldConnect, I could search on an ancestor and easily compare the different trees to see who had the most reliable and informative entries. Then I could download whole trees in a GEDcom file and import them into my own tree. I was using the (Freely Licensed Open Source Software) Gramps program on Linux, which made it very easy to fix GEDcom files and after importing, remove duplicates and fix inconsistencies. ( Gramps is also available for Windows and Mac. ) Within a year I had accumulated 700 direct ancestors in my tree. [see view of Gramps below] I posted my tree on WorldConnect linking back to my tree on my own website (http://quickening.zapto.org/gramps). The advantage of freely sharing your tree is that more people can access it, and they will find errors and tell you about them! It's like a self-correcting mechanism. The same can not be said about ancestry.com public member trees – it is too easy to replicate mistakes.
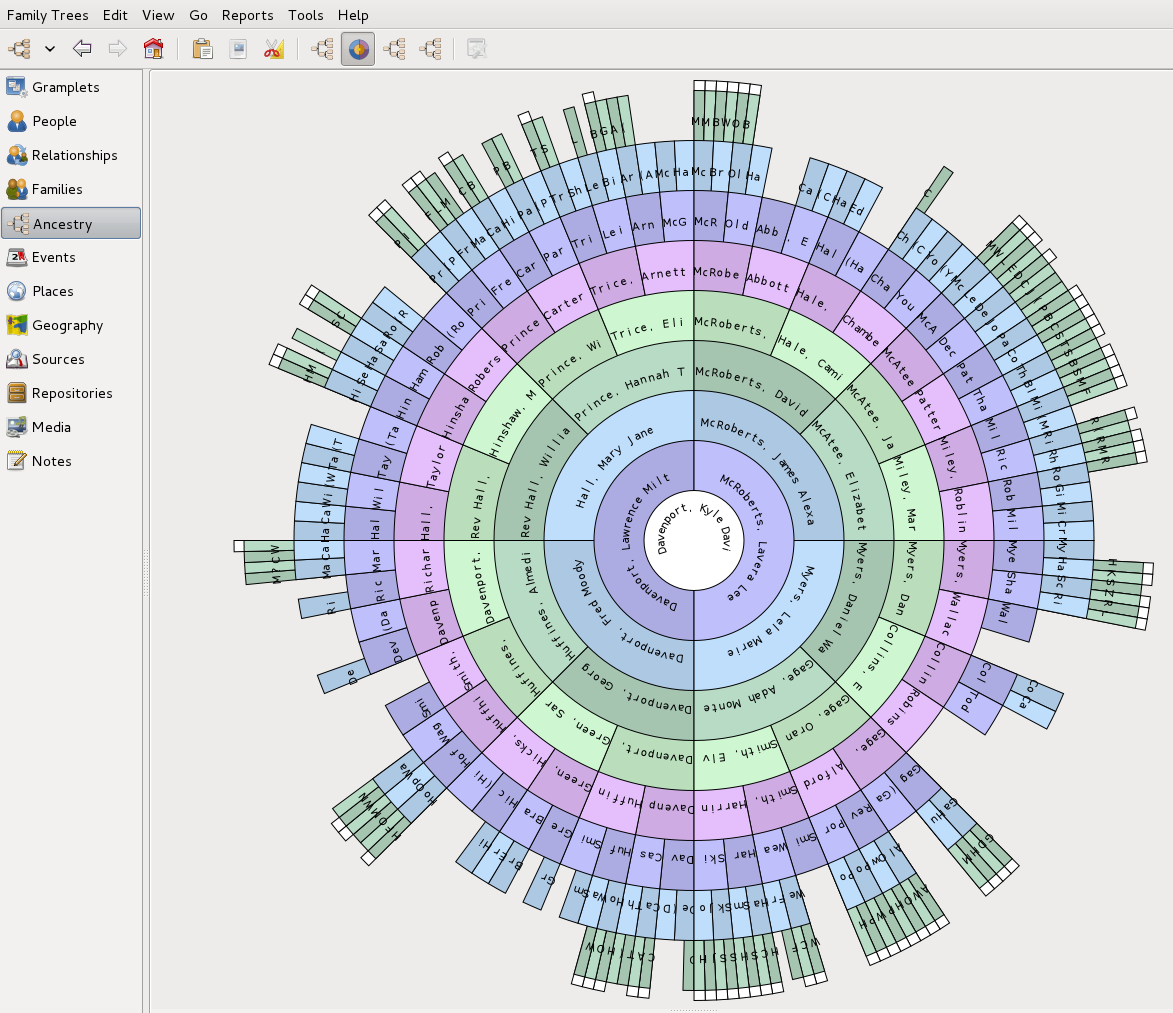
Importing trees of course is a gamble. You are depending on the other genealogist to have done their due diligence. As I worked back into the past, I would verify references and contact the researchers to reconcile inconsistencies. In another year, I was at a total of 1200 direct ancestors in my tree. And then it got hard! I would have to start doing some original research which meant buying death certificates, digging thru films at LDS centers, and building out my tree to include descendants of my ancestors – hoping I would come across someone or something to clue me in.
One of my earliest problems was where the Davenports came from. Generations of genealogists could trace my Davenports to The Four Brothers who arrived in Jackson Co, Tenn from Virginia in 1813, but no one could find any more detail. So my first massive project was to put all the Davenports of Jackson Co, TN in to my tree , and the families they married into, and their descendants – hoping some clue would pop out. But it didn't; I was still stuck. I noticed the given names of my Davenports were commonly used by another group of Davenports who originated in Virginia. Most Davenports in Virginia are in this group. They call themselves “Pamunkey Davenports” (Their patriarch was first found in Pamunkey Neck, Virginia). A new DNA test had just come out that these Pamunkey Davenports had taken, which was the only way I could prove I was related to them. The Y-DNA test could be taken only by men. Because men inherited the Y chromosome from their fathers along with his last name, it was also called a surname test.
I tested at Family Tree DNA and eagerly awaited the results. Five weeks later the results started trickling in. Not a Pamunkey! At 37 markers, I was an exact match to only 1 Davenport and 1 Eaton.
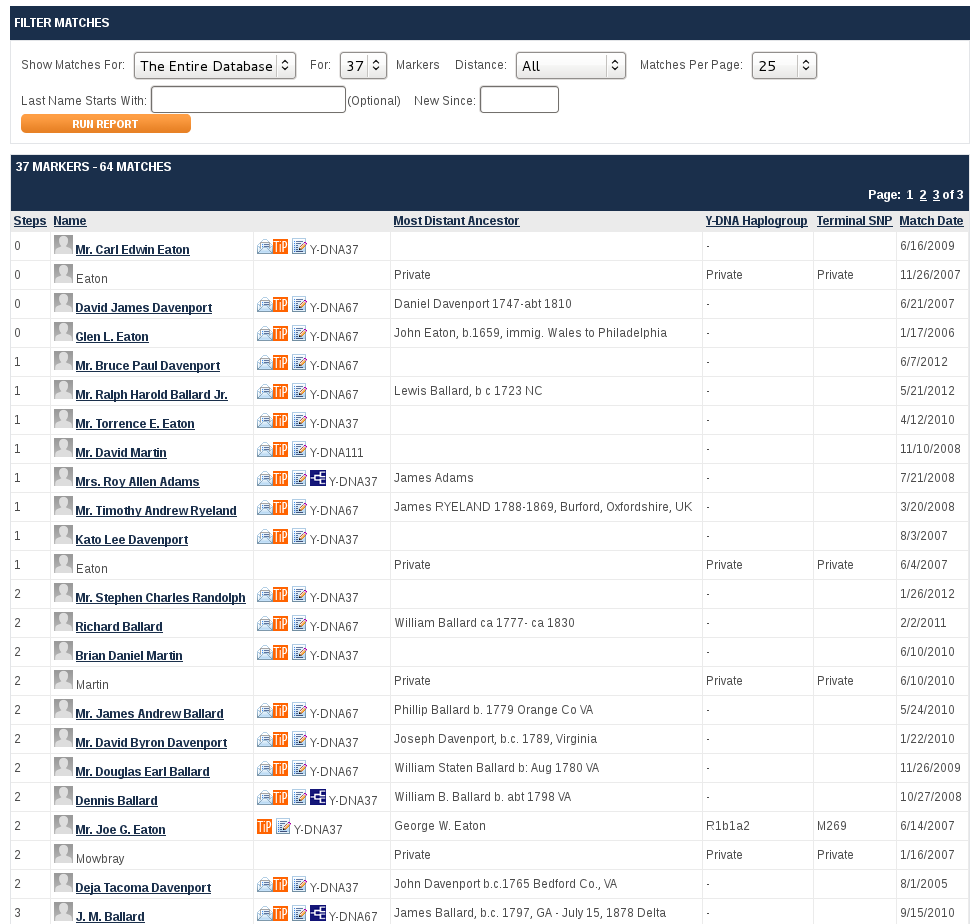
The Eaton researchers were really excited by that because of the famous association between John Davenport and Theophilus Eaton, founders of New Haven, CT. Unfortunately , we didn't match Eaton descendants of Theophilus or Davenport descendants of John. The other Davenport match also came from Jackson Co, Tenn and after my in-depth study of these Davenports, we established that he had come from a different one of the Four Brothers than I had. Both results convinced all 3 of us to upgrade to 67 markers. The Eaton was now 4 markers different from me, and a common ancestor was pushed back to 400+ years ago. The other Davenport and I were still exact matches in all markers , even though our common Davenport ancestor was 7 generations back.
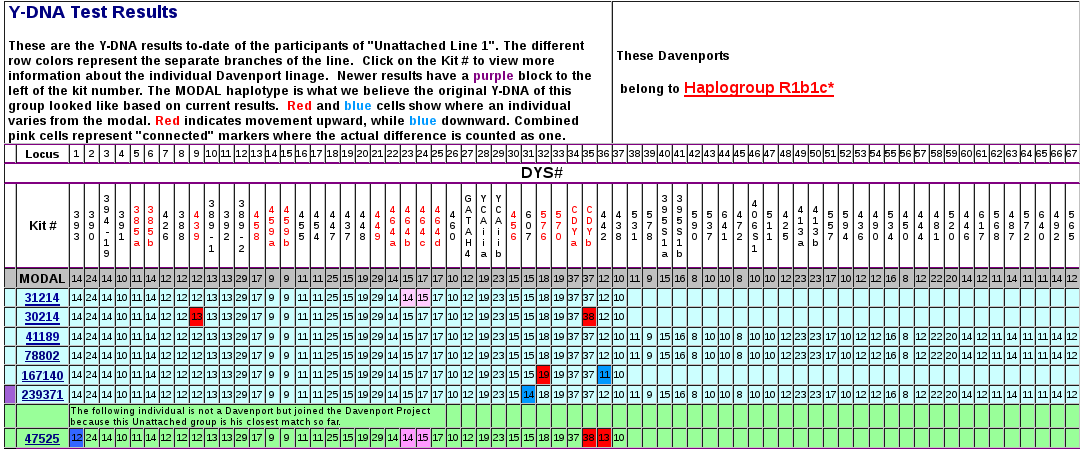
But how did this test help my genealogy? We Jackson Co, Tenn Davenports turned out to be close matches with 2 other Davenports: a descendant of John Davenport b.1767 Bedford Co, VA and a descendant of Thomas Davenport b.1823 KY. The first had 2 markers different, and the other just 1 (This is also callled the “Genetic Distance” or GD) . If you check the estimates of MRCA [Most Recent Common Ancestor] or matches with the same surname, and that means we are definitely related to them.
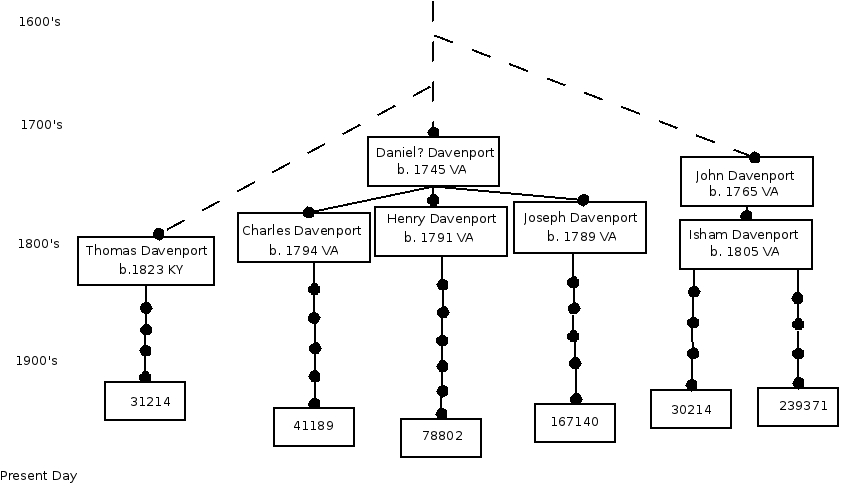
With an average likelihood of 1 mutation every 200 years, this result implies our Davenports go back much further than The Four Brothers of Jackson Co, Tenn. An exciting result indeed! The Davenport name of course is a well known line in Cheshire, England, but we don't match them. So we have to consider that an NPE [Non-Paternal Event] created the first Davenport in our line (even though he wasn't a Davenport). Other close matches we have would be good candidates for who that first Davenport was – Ballard, Eaton, Martin, Leigh... All definitely English.
A variety of coincidences lead us to suspect we descend from the Davenports of Prince George Co, Virginia. In fact, there are known descendants of these Davenports , but none have been tested yet – despite my best efforts to contact and convince them. Someday I hope they will be, and I would then be quite confident that that is where our Davenports came from – yes, even in the absence of a paper trail. In the meantime, more Davenports in our known lines have been tested, including a descendant of a 3rd brother in the original four. So 200 years later, DNA has proved the old family lore about the Four Brothers.
So it was, after accomplishing so much with the Y-DNA test, that I jumped on a DNA Day Sale at the genetic testing company 23andMe to get the new “DNA Chip” test which tests autosomal DNA – all the other chromosomes. The DNA chip or microarray works in an entirely different way than the Y-DNA test.
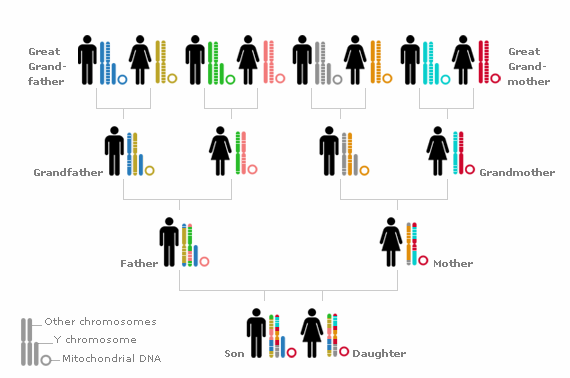
It identifies particular SNP's (Single Nucleotide Polymorphisms) - 300,000 to 1,000,000 of them across all 22 autosomal chromosomes and the 2 sex chromosomes. That means both men and women can find descendants of any of their ancestors, and not just the patrilineal or matrilineal ones. By testing so many markers , statistical methods can determine that matching sequences of SNP's probably mean a common ancestor. The biggest problem using autosomal markers is that pieces of the chromosomes are “shuffled around” in the next generation. The arms of each chromosome crossover 2 or 3 times at mostly random positions in every generation, and after 6 generations, only very small pieces from an ancestor are likely to be left. A common ancestor 6 generations back would be a 5th cousin.
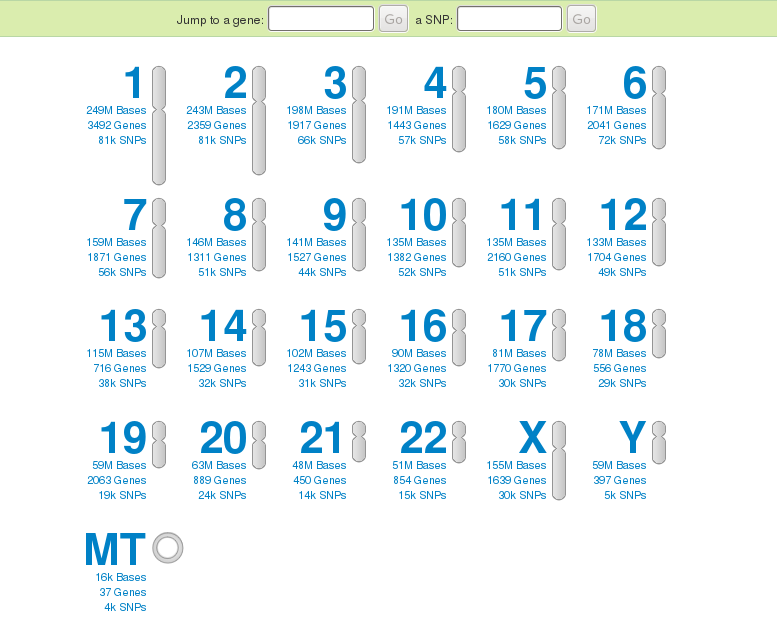
SNP's are chosen by geneticists who have determined which positions are most variable or affect gene expression. Currently about 10 million are known, so less than 1/10th of these SNP's are used in DNA chips which have the most significance for genes or for genealogy. Most of 23andMe's customers in fact get the test for health reasons, and do not care about matching relatives. They maintain and grow a long list of markers associated with particular traits or disease risks, good or bad. But what we are trying to match for relatedness are long strings of these markers. A string of 500 or more matching SNP's is considered significant. To put this in perspective, only about 1 in 2500 nucleotides are tested. So that string of 500 SNP's would actually be 1,250,000 base pairs (nucleotides) long. Geneticists use a different unit of measurement though to compare these lengths called the centiMorgan (cM) which is varying all along every chromosomes. It “normalizes” comparisons for relatedness purposes and represents how likely crossovers would occur in a region of the chromosome.
When the day arrived for my results , I logged into 23andMe with my kit#, and examined what I could do with my results. There were sections on Health, Ancestry, Sharing, and Research. Many of the SNP's tested are within genes and were chosen because they affect the function of the expressed protein. First of course, check out the disease risks... good, there was nothing serious. Then I looked at the traits... “breastfeeding and IQ”? Yes I had the gene associated with increased IQ if breastfed. Way to go, mom! I have “Bitter Taste Perception” - wow, that brings back memories of testing that in science class in 5th grade. Confirmed. Lots of interesting other traits – 57 in total. I don't see a “Likely to become a genealogist” trait listed – yet!
On to the Ancestry section. Maternal line confirmed, and some Y SNP's apparently identified my deep haplotype (ancestral origins of my paternal line), sparing me from taking other Deep Clade tests. Finally, click on Relative Finder.
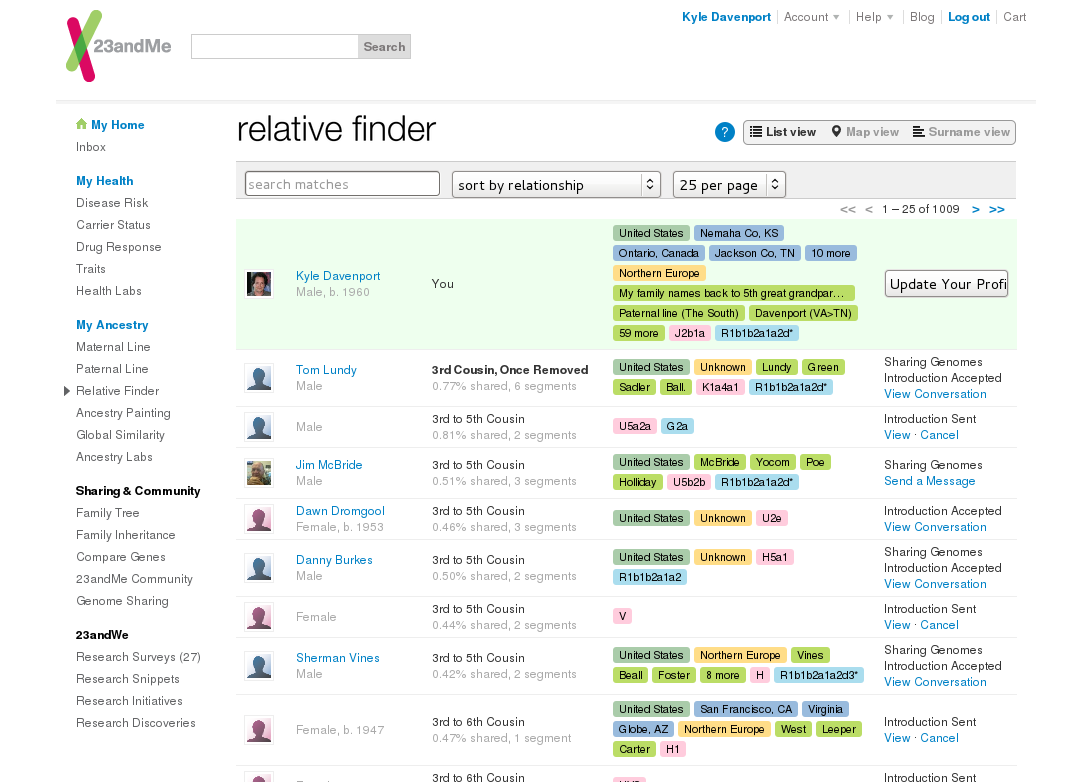
When I first visited Relative Finder , 2 years ago, I had less than 750 matches. Now it's up over 1000. Here are totals of the predicted relationships from my matches:
|
Predicted |
Count |
|---|---|
|
3rd Cousin |
4 |
|
4th Cousin |
170 |
|
5th Cousin |
818 |
|
Distant Cousin |
17 |
I set out to contact all my 3rd and 4th cousins back then, and also search for my surnames among the rest. They make it pretty easy to find interesting matches too – for instance, I can search on Marshall for any mention of the name in their profiles.
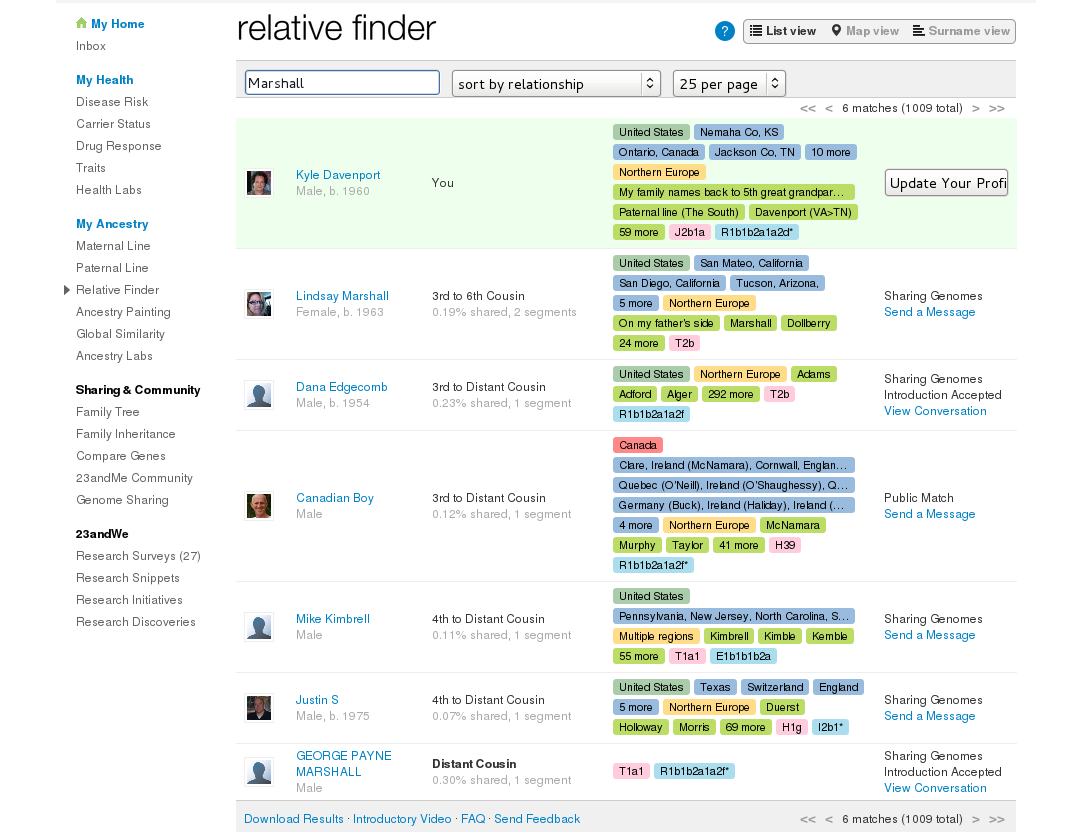
At this point however, I discovered most of my matches did not care about their ancestry:
|
Status |
Count |
|---|---|
|
Introduction Declined |
13 |
|
Introduction Ignored |
250 |
|
Introduction Accepted |
141 |
|
Public Match |
161 |
Of all the successful contacts, we reviewed our ancestry and if we found anything in common, like surnames or specific locations, we would then “Share Genomes”. I am “sharing genomes” with 80 of them. This allows you to compare your genome with theirs using 23andMe tools. One of these is “Compare Genes” which ranks your sharing matches by similar DNA and some traits (known genes).
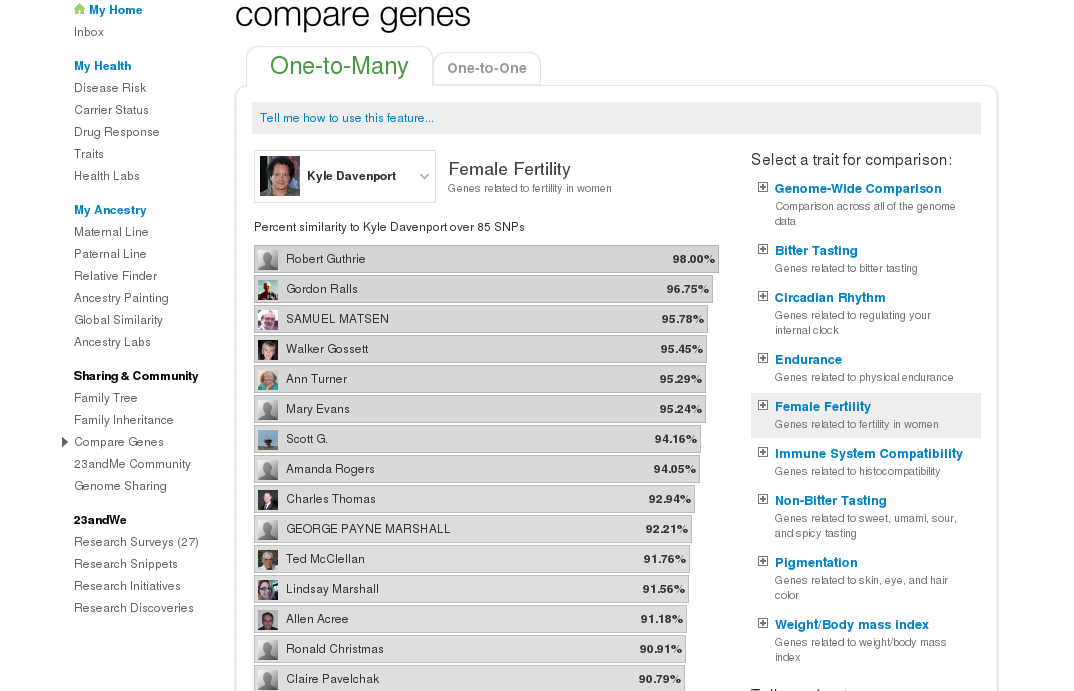
Payne and I are very similar in Female Fertility genes! Another tool is “Family Inheritance”, which shows a nice map of the chromosomes and where your matching segment lies.
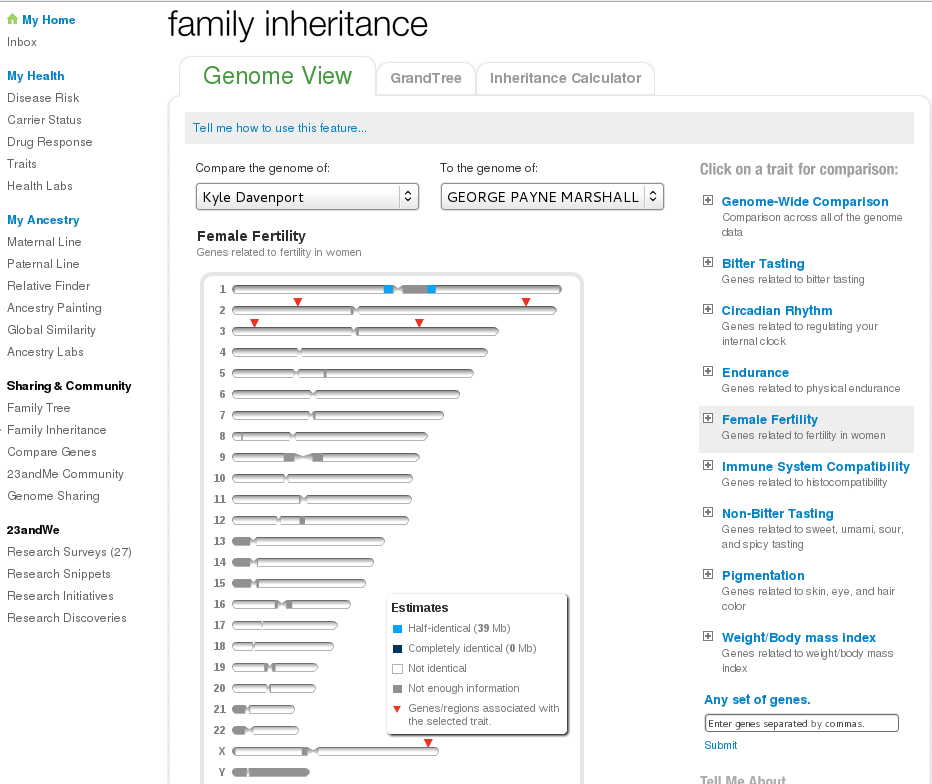
You can click on some traits and see if the genes for those traits lie within your matching segment. The fertility genes Payne and I share are not within our matching segment. An aside: many more genes and traits have been discovered than show up on 23andMe. Federal Dept of Energy maintains a “Gene Gateway” with nice maps of genes associated with disease.

I notice the Marshall Syndrome is just about where my matching segment with Payne is located!
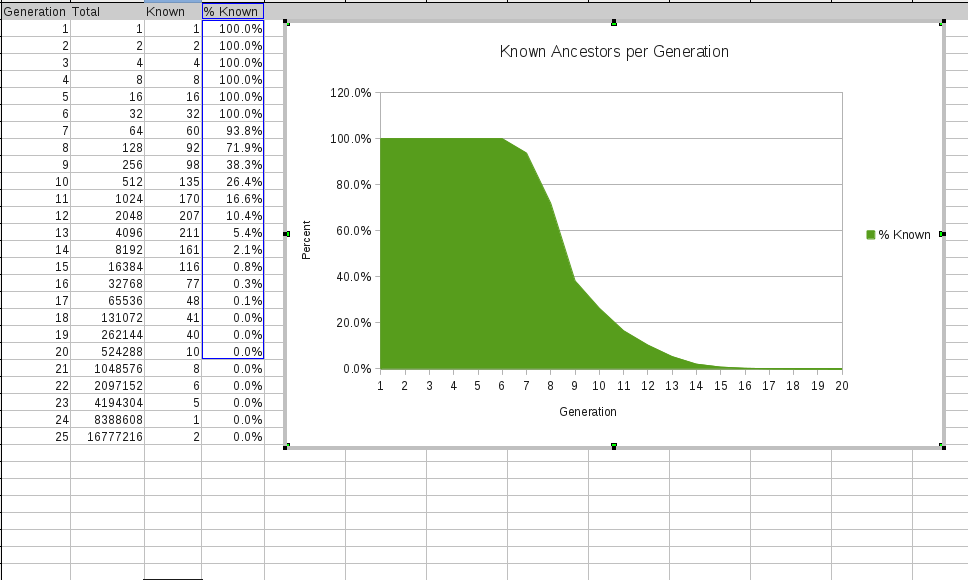
But out of all my matches who I shared with, I identified a common ancestor with only 10 of them. My closest match was a 3rd cousin-once removed whose parents I already had in my database. The other matches with known ancestors who were predicted 4th or 5th cousins, were more like 7th and 9th cousins! Not many people have trees going back 8 to 10 generations on all their lines, which seems to be about what is required to find common ancestors. Yes, somewhat disappointing, so I did a study of how many cousins I expected to find. With a liberal estimate of 1 million descendants from all my 4th great grandparents, and about 300,000 tested out of a US population of 300 million, I would expect 1 in a 1000 of my 5th cousins to have been tested – or about a 1000! Coincidentally , that is just how many matches I have. So perhaps if more people could complete their trees back that far, we really would be finding more common ancestors. It has certainly encouraged me to re-examine all the short branches on my tree; I now have over 1500 direct ancestors in there.
However, not all my cousins will show up as matches. The likelihood that a cousin will match drops off very quickly: only one half of 4th cousins and about one tenth of 5th cousins and less than 2% of 6th cousins are expected to show up as a match. [see Screenshot below]. So who are all those other matches? I believe it's what they call in statistics the “long tale”. Even though the fraction matching becomes very small, the number of my distant cousins is growing exponentially with each generation back. [E.g., twice the number of families * high fertility rate back then = 2 * 7. If I have 1 million 5th cousins , then I have about 14 million 6th cousins. ]
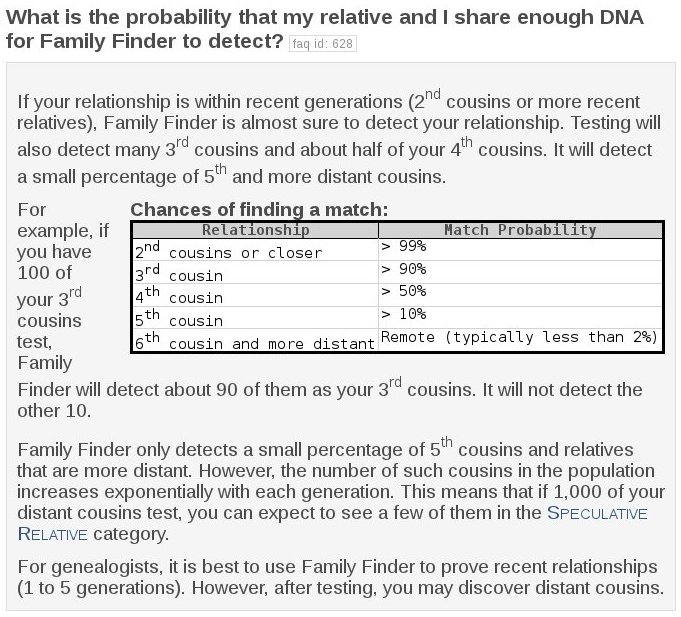
And, not all my matches are cousins! The reason 23andMe sets its matching threshold so high (7.5 cM) is to avoid “noise”. Matching segments can be IBD or IBS – Identical By Descent or Identical by State. By shear chance, and because we share a common ancestry far enough back, a shuffling of sections will result in an identical segment, rather like shuffling cards will eventually get you a straight flush. We are using statistical arguments to set these thresholds , and just like shuffling cards, at some point – a straight flush of 7 cards?! - it becomes really unlikely to have happened by chance. You are more likely to have inherited that straight flush from that new deck of cards you combined with the old deck, and it just hasn't been shuffled enough yet.
One more consideration is whether my matching segments are uniformly distributed along the chromosome.
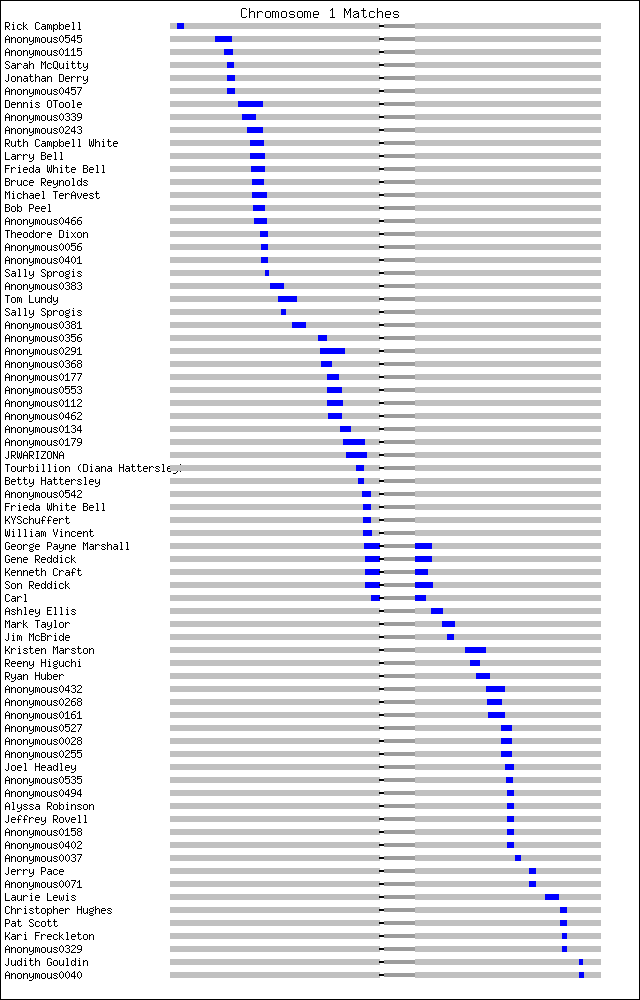
Clearly they are not! Even though the centiMorgan is supposed to incorporate the crossover frequency, you can see the matching segments are bunched up in two columns. This only happens on a few of the chromosomes, and other people have noticed many matches in the same ranges, but only graphically is it obvious. I have had no luck identifying common ancestry with any of the people in these ranges, and my best guess is that this region of the chromosome is conserved. I have either a higher number of IBS matches there or the common ancestor is much further back.
A problem trying to use autosomal DNA for genealogy is that a matching segment can come from any ancestor. Case in point, my match with Payne is predicted to be 4th cousin (shared 3rd great grandparents) , not my 8th great-grandparent Jacob Marshall! Although we are definitely distant cousins, it's unlikely the shared segment came from Jacob. How can we narrow down which ancestor that segment came from? In practice, the predicted relationships have been overly “generous”, and I find usually it must be 2 or more generations further back. So the first thing to do would be to compare our genealogies back to the 5th great grandparents at least. Not many of us have family trees that complete. I know fewer than 3/4 of my 5th great grandparents.
You can look through someone else's tree for common names and places, but that gets very tedious (especially for a thousand matches). Wouldn't it be great if the computer looked for us? I think ancestry.com attempts to do that, perhaps ftdna.com is also (not very well so I have heard). I know 23andMe has just started allowing trees for their members. In any case, this is how I found gedmatch.com. When you upload your GEDcom (hopefully an export from your genealogy software that includes only your relatives!) file to GEDmatch, you have some tools to look for matching names in other people's GEDcom files. I have found some distant cousins this way, but there are way too many false positives from incomplete information in some people's trees! While on GEDmatch I discovered all the other tools for comparing DNA, especially with tests from other companies. More on that later.
I have some matches that I share several ancestors with. How can you tell which ancestors the DNA segment came from? The easiest way to exclude half your tree is to have your results phased. The raw results are hundreds of thousands of pairs of nucleotides (A,G,T, or C) (1 from each parent; 1 on each arm of the chromosome). The nature of the DNA probe used in the test can not determine which arm, or which parent, a particular allele came from. But if you phase your results, they will be lined up in the proper sequence. You might think you can only phase your results by comparing them to a parent or very close cousin, but in fact, statistical methods based on common ancestral orderings can also give a good guess. [see Beagle, a work in progress] I understand that ancestry.com does this automatically before attempting any matches. Although of course, for a single test result you will not know which phase is maternal or paternal.
The other way, if you are very lucky, is to have a third match with approximately the same shared segment. If at least one of the endpoints of the segment are approximately the same, and your 2nd and 3rd matches also match each other, than you probably all got the segment from a common ancestor. This allows you to triangulate on that common ancestor (and exclude the others). In practice of course few people can find a single common ancestor beyond close cousins, but in principle, this could help genealogists. If your 2 other matches have in common the same specific location that you do ( a particular county in some state, for example), then it could be a clue that you are related to the people they knew about in those locations. Knowing those other surnames could give research hints and narrow down search results. [again see Kyle_Matches_Chromosome1 above] Case in point, see Gene Reddick and Payne sharing same segment with me. Gene has a fairly extensive tree with some tantalizing commonalities, but no specific names. But we both seem to share Rhea's in Augusta Co, VA, and the Marshall's were there too.
What else can you do with your results? Many people are more interested in their deep ancestry, aka, am I part-Indian? If you take the “Where are you From?” survey on 23andMe , then you go in to their Ancestry Labs menu and look at your Ancestry Finder tool.
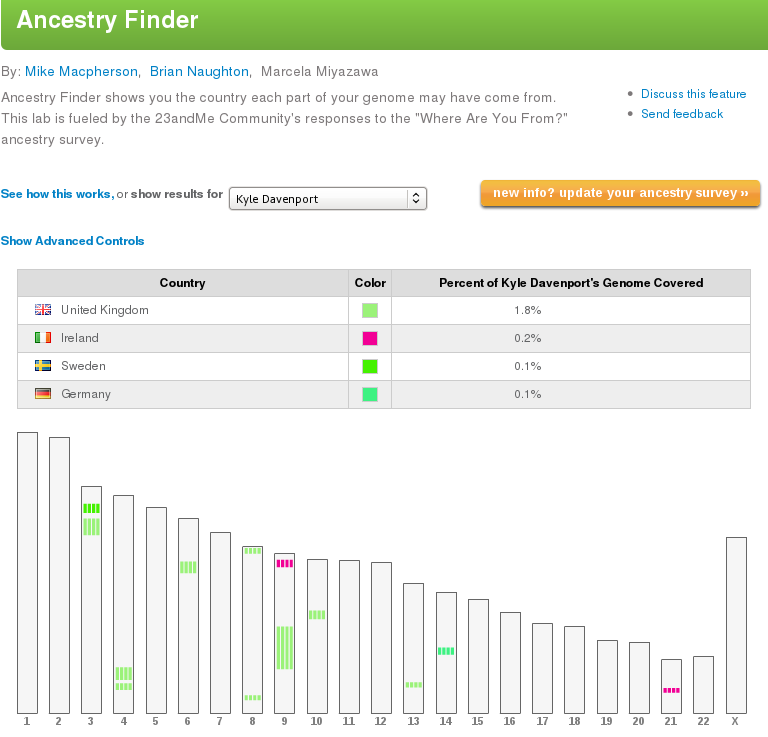
I'm mostly English, about 1/4 Irish and some Scottish. But on this page you can download all your matches – those who have taken the survey – into a nice spreadsheet. I highly recommend you do this! It shows the exact locations and size of your matching segments. I add to this all the other matches I am sharing with, and matches I have from GEDmatch.com. My chromobrowse program used this data to display the matching segments. For shared genome matches who haven't taken the survey, you can find the exact numbers in Ancestry Labs->Family Inheritance:Advanced. There's also a Haplogroup Mapper which identifies the mutations they used to assign your haplogroup. This can be fascinating when you search on these mutations to find the deep origins of your paternal or maternal ancestor. Mine (U152) traces back through the Urnfield, Hallstatt, La Tene Celt and Bell Beaker cultures of central Europe. This mutation probably originated 10,000 years ago. A smaller fraction of U152 have DYS492=14 is more common in southern Germany.
Ancestry.com and FTDNA.com are other companies offering an autosomal test for genealogy. They are probably more genealogy oriented than 23andMe. Ancestry.com would be ideal with their experience in family trees, but currently they are still in beta. How can you compare your results on 23andMe to people who tested on FamilyTreeDNA? By uploading your raw results to GEDmatch.com. It's a free service done by enthusiastic amateurs who have incorporated an amazing variety of tools to analyze both GEDcoms and DNA. At least, your matches on GEDmatch will be serious about genealogy.
GEDmatch's tools are better for finding matches for several reasons. 1) You can change the thresholds for matches either in SNP's or cM's When I expect a match between 2 people I will lower the cM to 3. 2) GEDmatch will still identify a match even if there are a few mutations with the matching segment. This seems entirely reasonable to me, that in a stretch of thousands of matching SNP's there might be a few mutations. 3) It color codes the matching SNPs. One might expect a distant cousin to have many smaller matching segments, even if there are none above threshold. What you see in the displayed chromosome map is lot more green than red. 4) Another tool will find overlapping segments, allowing you to triangulate on an ancestor. 5) The matching tool allows you to pick any 2 tests of other people so you can see if the overlapping segment is the same between them. 6) You can compare GEDcoms with your matches. 7) You can triangulate your matches ( find people in your matches who also match each other) For a good comparison of the color coding, check 2 close matches (same cM) , but the greener one I have identified multiple common ancestors with, while the other one is mixed African-American.
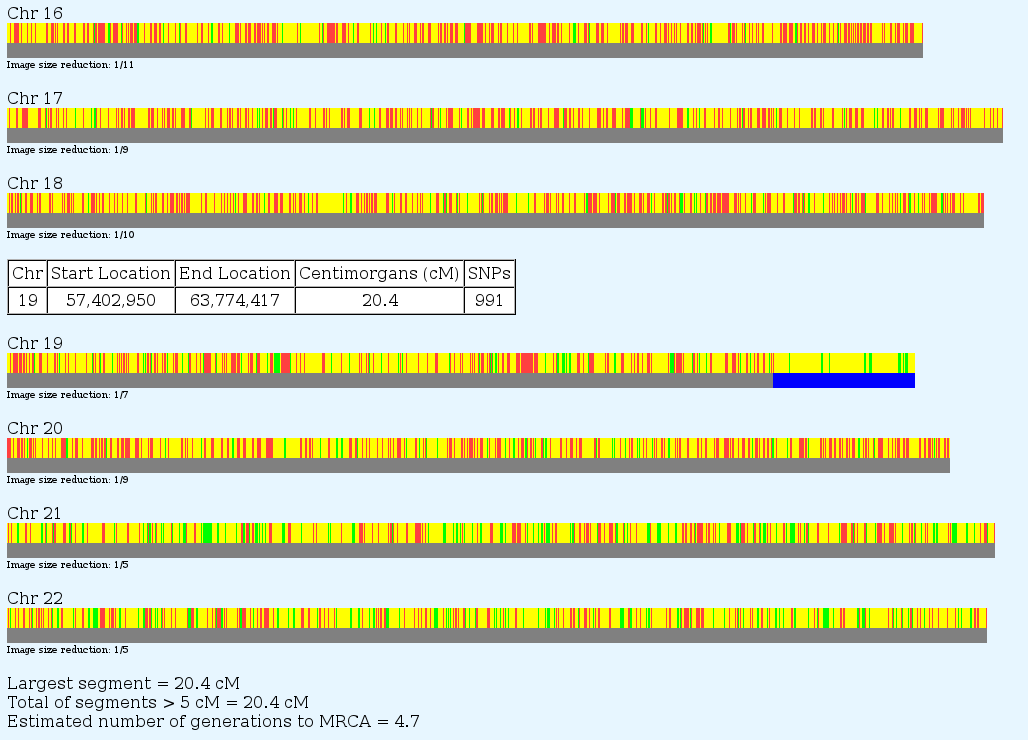
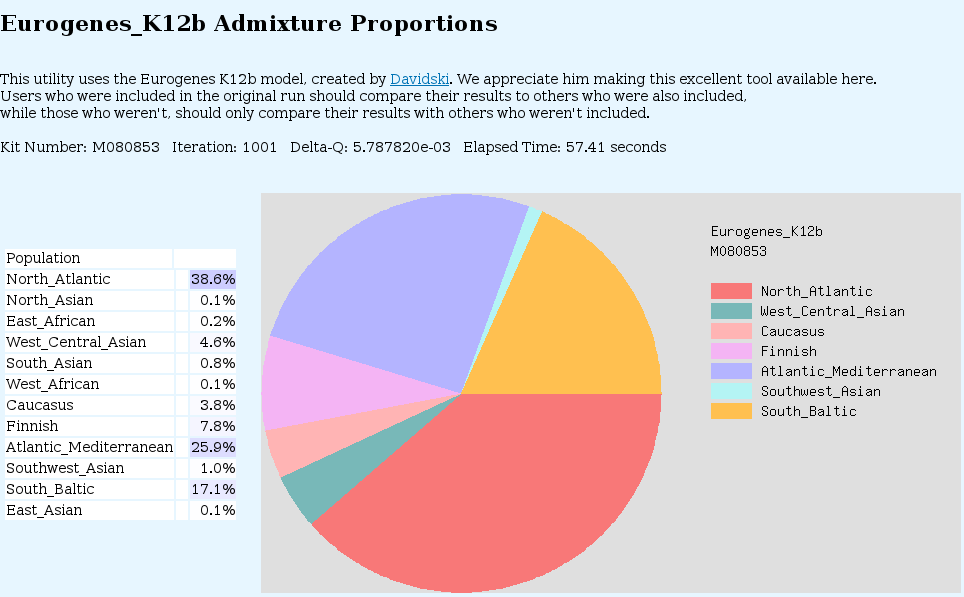
One utility on GEDmatch not to be missed is Admixture-Proportions Analysis. Unlike the deep ancestry haplotype tests which test only your paternal or maternal line, AdMix tests are statistical inferences based on certain rare SNP's (Ancestry Informative Markers) and their likelihood of appearing in various populations. There are a variety of models and different ways of displaying the results. See below:
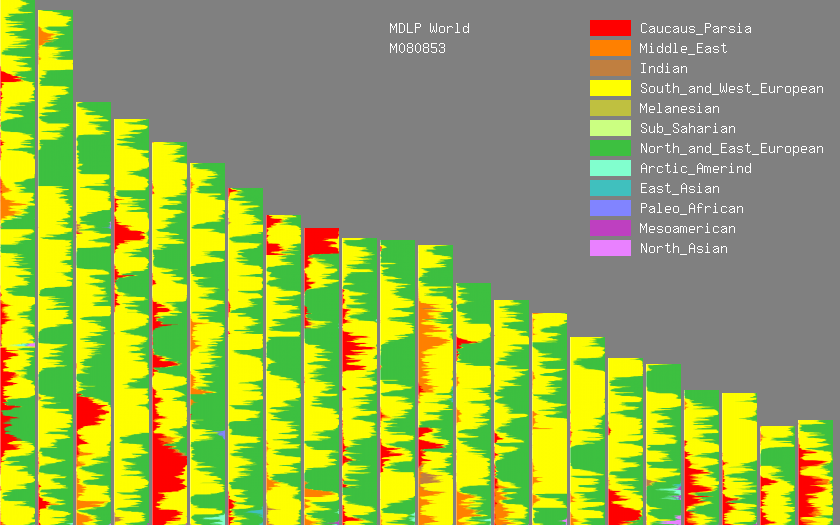
You can zoom in for more detail
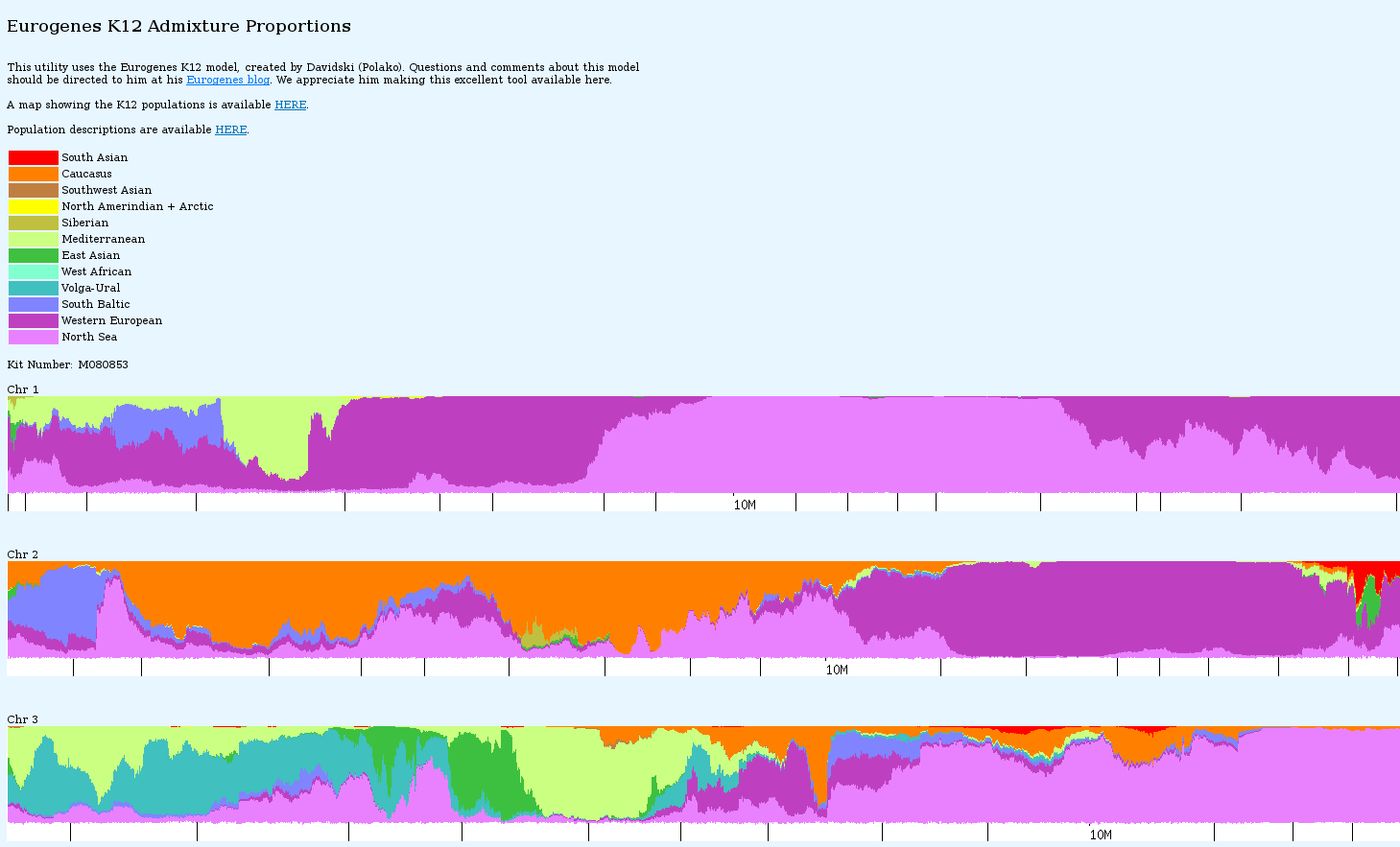
See way down on chromosome 18 there is a patch of yellow – NorthAmerindian . Yes, I probably have some Indian in me 10 generations back.
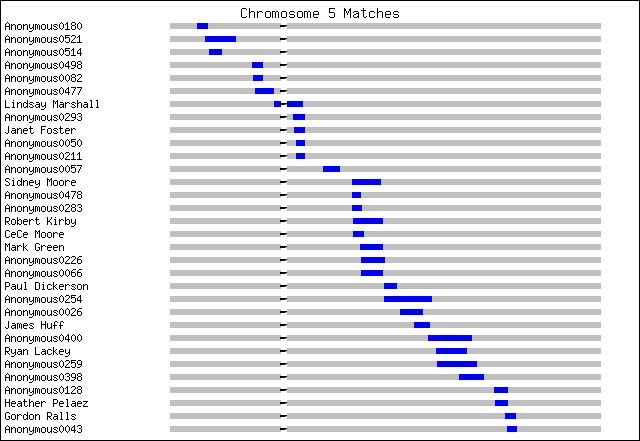
The alternative of course, is to test at both 23andMe and FTDNA. For a small fee you can also transfer your recent results from 23andMe to FTDNA. The majority of genetic genealogists have come to expect not much from autosomal tests beyond 2nd cousins, perhaps 3rd – at this time! They think it is more useful to have lots of close relatives tested, then to guess how more remote cousins are related. Testing close relatives on your maternal or paternal side can let you phase your results. [see Kyle_Matches_Chromosome5.png; note Sidney Moore is CeCe Moore's paternal uncle]
Testing a sibling is good for doubling the number of your “matches”; you share on average 50% of your DNA with another sibling but 100% certainty a match to her is related to you (even if there are no matching segments between you and her match). I had my sister tested at FTDNA., and found many more matches – especially among genealogists! Similarly, any close relative will share much more DNA with you then the typical match
When the prices fall and the tests get more comprehensive, and many, many more get tested, there will definitely be new opportunities for statistical analysis.
My impression from comparing trees with my matches is that I should be finding more common ancestors. Especially among my close matches at 3rd and 4th cousin estimation, both of us would have fairly complete trees back 8 generations and still find no common surnames! Genetic genealogy has shown that, conservatively, 1.9% of each generation is a Non-Paternal Event. The 1.9% is a lower bound based on studies of “solid paper trail” lineages. Y-DNA project admins are reporting 5 to 10 % of those tested for paternal lineage turn out to be NPE's. If you add up all these generations going back along all your lines, clearly you get a very high proportion of wrong branches in your tree. At the same time genetics is showing infidelities and bad genealogies, it is reconnecting adoptees with their biological families. Many of my matches got the test just because they or their parents or grandparents were adopted ( or disappeared , or changed their names...) Our real trees are probably very “messy”... [ The stories I could tell! ]
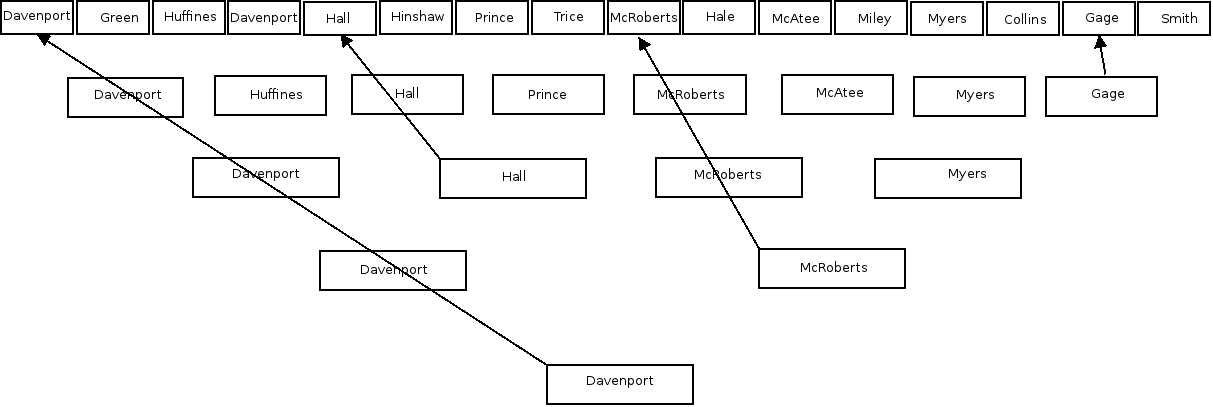
Y-DNA tests can prove there was no NPE at least as far back as the Most Recent Common Ancestor (the surname patriarch). But what about the rest of your tree? [see Kyle-Surname-Tree.png] Use Y-DNA tests on them too! Find male cousins who are patrilineal descendants along all your lines and see if they match others with the same name – hopefully very far back. I've done this with many of my lines with varying success. My mom was a McRoberts and I tested a cousin McRoberts. We found matches to other McRoberts where the common ancestor must be greater than 8 generations back. Unfortunately when I tested a male Hall, cousin of my dad's mom, no other Hall matched him. Similarly, testing a male Smith, distant cousin of my gggrandmother, no other Smith matched him either.
What to Expect in the Future
More SNP's; more FGS's (Full Genome Sequencing)
Automatic phasing
Cheaper tests which will allow many more to test
More ancient DNA
New Genealogy oriented chips like Geno 2.0
Geno 2.0 is a new chip developed by National Geographic Society specifically to address ancestral origins. It will test about 150,000 AIM's (Ancestry Informative Markers) which have only recently been identified by various DNA Atlases, Walk The Y, and FGS (Full Genome Sequencing). They deliberately excluded SNP's within genes or SNP's associated with medical conditions. By emphasizing a huge number of rare SNP's, they predict they will be able to identify ancestral origins up to 1000 years ago. That is, they will fill in the gap between Relative Finder and Haplotypes. It obsoletes current Deep Clade tests because it will have 12,000+ Y SNP's. In Beta – expect testing to start Oct 30th.
Your Genetic Genealogist describes Geno 2.0
test available initially only from National Geographic Society , but will be offered by FTDNA:
National Geographics Geno 2.0 site.
More Resources
"DNA Research for Genealogists: Beyond the Basics"
Genetic Genealogist professional, her blog (and one of my matches ;):
Roberta Estes site - nice introductory articles.
Autosomal DNA tools:
Mailing Lists:
Various other genetic genealogy lists
Favorite Free Genealogy sites:
Infoweb at Newsbank (obituaries & old newspapers; free with library card)
FamilySearch / LDS
Google! / Archives.org / Google Books